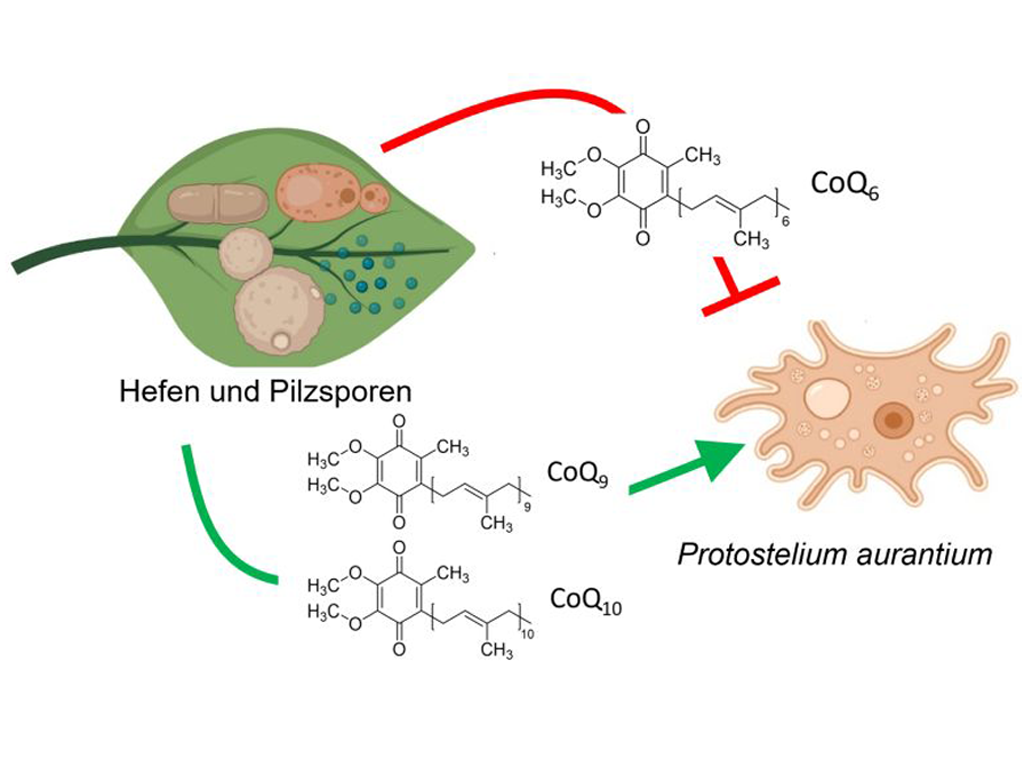
Fungi are major players in terrestrial ecosystems, with only some of them causing infections in higher organisms including humans. Mutual interactions with other microorganisms are among the key factors for their diversity, but are also increasingly recognized as a driving force for virulence. Among these, predation is known to cause substantial selective effects in higher organisms, summarized as the evolutionary arms race, but little is known how such relationships have shaped microbial communities. The Amoebozoa form a highly diverse group within the eukaryotic kingdom comprising mostly unicellular microorganism which all share a predatory lifestyle. Their undefined cell shape allows them to enclose and incorporate microbial cells by phagocytosis, a process that is also exploited by innate immune cells to eradicate invading bacteria and fungi. Despite their role as environmental predators and potential to study microbial phagocytosis, their complex biology has limited the number of model amoebae to a single one – Dictyostelium discoideum.
Within this project we will exploit P. aurantium, a recently isolated fungivorous amoeba, to study the selective effects and molecular targets of phagocytic predation on fungal cells. A fully annotated genome and established protocols for large scale cultivations with different fungal prey species, including human pathogens, have made P. aurantium a suitable model to study fungal recognition and killing in a high throughput manner. Using a targeted approach to elucidate the role of essential cofactors, in combination with a large scale screening of defined mutant libraries, the project aims to identify single genes and metabolic pathways that have been under selective pressure by environmental phagocytes. As the process of phagocytosis is highly conserved from amoebozoa to innate immune cells, it is well conceivable that some of them will prove to be highly relevant during infection of higher hosts. The project is meant to open a rarely considered ecological and evolutionary perspective to fungal virulence and may also identify new targets for antifungal drugs and diagnostics.